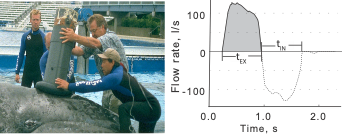
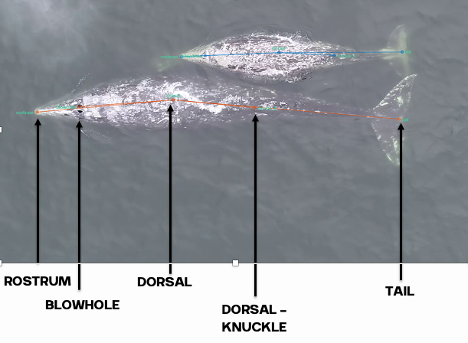
Clara Bird, PhD Candidate, OSU Department of Fisheries, Wildlife, and Conservation Sciences, Geospatial Ecology of Marine Megafauna Lab
Graduate school is an odd phase of life, at least in my experience. You spend years hyperfocused on a project, learning countless new skills – and the journey is completely unique to you. Unlike high school or undergrad, you are on your own timeline. While you may have peers on similar timelines, at the end of day your major deadlines and milestone dates are your own. This has struck me throughout my time in grad school, and I’ve been thinking about it a lot lately as I approach my biggest, and final milestone – defending my PhD!
I defend in just about two months, and to be honest, it’s very odd approaching a milestone like this alone. In high school and college, you count down to the end together. The feelings of anticipation, stress, excitement, and anticipatory grief that can accompany the lead-up to graduation are typically shared. This time, as I’m in an intense final push to the end while processing these emotions, most of the people around me are on their own unique timeline. At times grad school can feel quite lonely, but this journey would have been impossible without an incredible community of people.



A central contradiction of being a grad student is that your research is your own, but you need a variety of communities to successfully complete it. Your community of formal advisors, including your advisor and committee members, guide you along the way and provide feedback. Professors help you fill specific knowledge and skill gaps, while lab mates provide invaluable peer mentorship. Finally, fellow grad students share the experience and can celebrate and commiserate with you. I’ve also had the incredible fortune of having the community of the GRANITE team, and I’ve recently been reflecting on how special the experience has been.
To briefly recap, GRANITE stands for Gray whale Response to Ambient Noise Informed by Technology and Ecology (read this blog to learn more). This project is one of the GEMM lab’s long-running gray whale projects focused on studying gray whale behavior, physiology, and health to understand how whales respond to ocean noise. Given the many questions under this project, it takes a team of researchers to accomplish our goals. I have learned so much from being on the team. While we spend most of the year working on our own components, we have annual meetings that are always a highlight of the year. Our team is made up of ecologists, physiologists, and statisticians with backgrounds across a range of taxa and methodologies. These meetings are an incredible time to watch, and participate in, scientific collaboration in action. I have learned so much from watching experts critically think about questions and draw inspiration from their knowledge bases. It’s been a multi-year masterclass and a critically important piece of my PhD.

These annual meetings have also served as markers of the passage of time. It’s been fascinating to observe how our discussions, questions, and ideas have evolved as the project progressed. In the early years, our presentations shared proposed research and our conversations focused on working out how on earth we were going to tackle the big questions we were posing. In parallel, it was so helpful to work out how I was going to accomplish my proposed PhD questions as part of this larger group effort. During the middle years, it was fun to hear progress updates and to learn from watching others go through their process too. In grad school, it’s easy to feel like your setbacks and stumbles are failures that reflect your own incompetence, but working alongside and learning from these scientists has helped remind me that setbacks and stumbles are just part of the process. Now, in the final phase, as results abound, it feels extra exciting to celebrate with this team that has watched the work, and me grow, from the beginning.

We just wrapped up our last team meeting of the GRANITE project, and this year provided a learning experience in a phase of science that isn’t often emphasized in grad school. For graduate students, our work tends to end when we graduate. While we certainly think about follow-up questions to our studies, we rarely get the opportunity to follow through. In our final exams, we are often asked to think of next steps outside the constraints of funding or practicality, as a critical thinking exercise. But it’s a different skillset to dream up follow-up questions, and to then assess which of those questions are feasible and could come together to form a proposal. This last meeting felt like a cool full-story moment. From our earliest meetings determining how to answer our new questions, to now deciding what the next new questions are, I have learned countless lessons from watching this team operate.

There are a few overarching lessons I’ll take with me. First and foremost, the value of patience and kindness. As a young scientist stumbling up the learning curve of many skills all at once, I am so grateful for the patience and kindness I’ve been shown. Second, to keep an open mind and to draw inspiration from anything and everything. Studying whales is hard, and we often need to take ideas from studies on other animals. Which brings me to my third takeaway, to collaborate with scientists from a wide range of backgrounds who can combine their knowledges bases with yours, to generate better research questions and approaches to answering them.
I am so grateful to have worked with this team during my final sprint to the finish. Despite the pressure of the end nearing, I’m enjoying moments to reflect and be grateful. I am grateful for my teachers and peers and friends. And I can’t wait to share this project with everyone.




P.S. Interested in tuning into my defense seminar? Keep an eye on the GEMM lab Instagram (@gemm_lab) for the details and zoom link.
Did you enjoy this blog? Want to learn more about marine life, research, and conservation? Subscribe to our blog and get a weekly alert when we make a new post! Just add your name into the subscribe box below!