Puffy snout syndrome: though it has a cute-sounding name, this debilitating condition causes masses on the face of Scombridae fish (a group of fish that includes mackerel and tuna.) Fish afflicted with puffy snout syndrome (PSS) develop excessive collagenous tumor-like growths around the eyes, snout, and mouth. This ultimately leads to visual impairment, difficulty feeding, and eventual death. PSS is surprisingly confined to just fish raised in captivity – those in aquaculture farms or aquariums, for example. Unfortunately, when PSS is identified in aquaculture, the only option is to cull the entire tank — no treatments or cures currently exist.

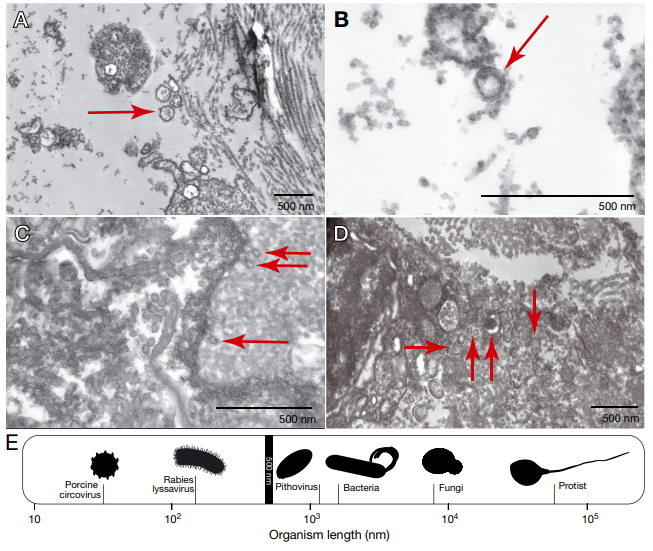
PSS was first identified in the 1950s, in a fish research center in Honolulu, Hawaii. Since then, there have only been 9 publications in the scientific literature documenting the condition and possible causes, although the fish community has come to the conclusion that PSS is likely a transmittable condition with an infectious agent as the cause. But despite this conclusion, there’s been no success so far in identifying such a cause – tests for parasites, bacterial growth, and viruses have come up empty-handed. That was until a 2021 paper, using high-resolution electron microscopy, found evidence of viral particles in facial tissues taken from Pacific mackerel. Suddenly, there was a lead: could PSS be caused by a virus that we just don’t have a test for yet?
Putting Together the Pieces
To investigate this hypothesis, this week’s guest Savanah Leidholt (a co-author of the 2021 microscopy study) is using an approach for viral detection known as metatranscriptomics. Leidholt, a fourth year PhD candidate in the Microbiology department, sees this complex approach as a sort of puzzle: “Your sample of RNA has, say, 10 giant jigsaw puzzles in it. But the individual puzzles might not be complete, and the pieces might fit into multiple places, so your job is to reassemble the pieces into the puzzles in a way that gives you a better picture of your story.”

RNA, or ribonucleic acid, is a nucleic acid similar to DNA found in all living organisms, But where DNA is like a blueprint – providing the code that makes you, you; RNA is more like the assembly manual. When a gene is expressed (meaning the corresponding protein is manufactured), the double-stranded DNA is unwound and the information is transcribed into a molecule called messenger RNA. This single-stranded mRNA is now a copy of the gene that can be translated into protein. The process of writing an mRNA copy of the DNA blueprint is called transcription, and these mRNA molecules are the target of this metatranscriptomics approach, with the prefix “meta” meaning all of the RNA in a sample (both the fish RNA and the potential viral RNA, in this case) and the suffix “omics” just referring to the fact that this approach happens on a large scale (ALL of the RNA, not just a single gene, is sequenced here!) When mRNA is sequenced in this manner, the researchers can then conclude that the gene it corresponds to was being expressed in the fish at the time the sample was collected.

So far, Leidholt has identified some specific genes in fish that tend to be much more abundant in fish from captive settings versus those found in the wild. Could these genes be related to why PSS is only seen in fish in captivity? It’s likely – the genes identified are immune markers, and the upregulation of immune markers is well-known to be associated with chronic stress. Think about a college student during finals week – stress is high after a long semester, maybe they’ve been studying until late in the night and not eating or sleeping well, consuming more alcohol than is recommended. And then suddenly, on the day of the test, they’re stuck in bed with the flu or a cold. The same thing can happen to fish (well, maybe not the part where they take a test!,) especially in captivity – Pacific mackerel, tuna, and other scombrid species susceptible to PSS are fairly large, sometimes swimming hundreds of miles in a single day in the ocean. But in captivity, they are often in very small tanks, constantly swimming in constrained circles. They’re not exposed to the same diversity of other fish, plankton, prey, and landscape as they would be in the wild. “Captivity is a great place to be if you’re a pathogen, but not great if you’re a fish”, says Leidholt.
The results of Leidholt’s study are an exciting step forward in the field of PSS research, as one of the biggest challenges currently facing aquaculture farms and aquariums is that there is no way to screen for PSS in healthy fish before symptoms begin to show. Finding these marker genes that appear in fish that could later on develop PSS means that in the future a test could be developed. If vulnerable fish could be identified and removed from the population before they begin to show symptoms and spread the condition, then it would mean fish farmers no longer have to cull the entire tank when PSS is noticed.
The elusive virus
One of the challenges that remains is going beyond the identification of genes in the fish and beginning to identify viruses in the samples. Viruses, which are small entities made up of a DNA or RNA core and a protective protein coating, are thought to be the most abundant biological entities on the planet Earth – and the smallest in terms of size. They usually get a bit of a bad reputation due to their association with diseases in humans and other animals, but there are also viruses that play important positive roles in their ecosystems – bacteriophages, for example, are viruses that infect bacteria. In humans, bacteriophages can attack and invade pathogenic or antibiotic-resistance bacteria like E. coli or S. aureus (for more information on phages and how they are actually studied as a potential therapy for infections, check out this November 2021 interview with Miriam Lipton!) Across the entire planet there are estimated to be between 10^7 to 10^9 distinct viral species – that’s between 10 million and 10 billion different species. And fish are thought to host more viruses than any other vertebrate species. Because of technological advancements, these viral species have only really been identified very recently, and identification still poses a significant challenge.
As a group, viruses are very diverse, so one of the challenges is finding a reliable way to identify them in a given sample. For bacteria, researchers can use a marker gene called the 16S rRNA gene – this gene is found in every single bacterial cell, making it universal, but it also has a region of variability. This region of variability allows for identification of different strains of bacteria. “Nothing like 16S exists for viruses,” Leidholt says. “Intense sequencing methods have to be used to capture them in a given sample.” The metatranscriptomic methods that Leidholt is using should allow her to capture elusive viruses by taking a scorched earth approach – targeting and sequencing any little bit of RNA in the sample at all, and trying to match up that RNA to a virus.
To learn more about Savanah’s research on puffy snout syndrome, her journey to Oregon State, and the amazing outreach she’s doing with high school students in the Microbiology Department, tune in to Inspiration Dissemination on Sunday, November 20th at 7 PM Pacific!
