Basic biology and computer science is probably not an intuitive pairing to think of, when we think of pairs of scientific disciplines. Not as intuitive as say biology and chemistry (often referred to as biochem). However, for Joseph Valencia, a third year PhD student at OSU, the bridge between these two disciplines is a view of life at the molecular scale as a computational process in which cells store, transmit, and interpret the information necessary for survival.
Think back to your 9th or 10th grade biology class content and you will (probably? maybe?) vaguely remember learning about DNA, RNA, proteins, and ribosomes, and much more. In case your memory is a little foggy, here is a short (and very simplified) recap of the basic biology. DNA is the information storage component of cells. RNA, which is the focus of Joseph’s research, is the messenger that carries information from DNA to control the synthesis of proteins. This process is called translation and ribosomes are required to carry out this process. Ribosomes are complex molecular machines and many of them can also be found in each of our cells. Their job is to interpret the RNA. The way this works is that they attach themselves to the RNA, they take the transcript of information that the RNA contains, interpret it and produce a protein. The proteins fold into a specific 3D shape and the shape determines the protein’s function. What do proteins do? Basically control everything in our bodies! Proteins make enzymes which control everything from muscle repair to eye twitching. The amazing thing about this process is that it is not specific to humans, but is a fundamental part of basic biology that occurs in basically every living thing!
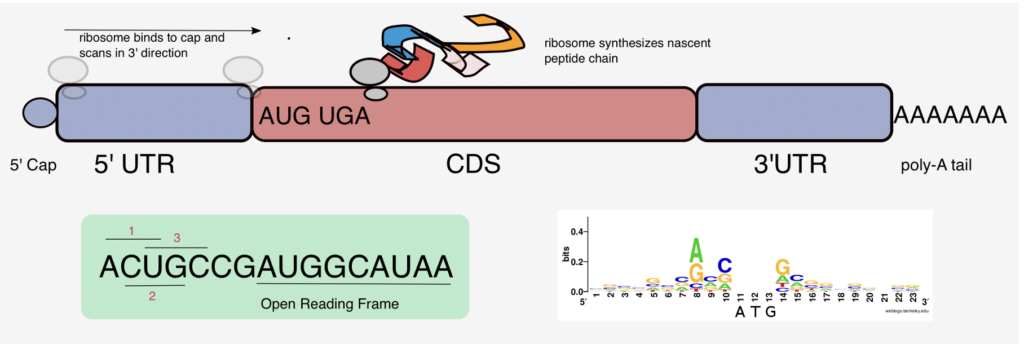
So now that you are refreshed on your high school biology, let us tie all of these ‘basics’ to what Joseph does for his research. Joseph’s research focuses on RNA, which can be broken down into two main groups: messenger RNA (mRNA) and non-coding RNA. mRNA is what ends up turning into a protein following the translation by a ribosome, whereas with long non-coding RNA, the ribosome decides not to turn it into a protein. While we are able to distinguish between the two types of RNA, we do not fully understand how a ribosome decides to turn one RNA (aka mRNA) into a protein, and not another (aka long non-coding RNA). That’s where Joseph and computer science come in – Joseph is building a machine learning model to try and better understand this ribosomal decision-making process.
Machine learning, a field within artificial intelligence, can be defined as any approach that creates an algorithm or model by using data rather than programmer specified rules. Lots of data. Modern machine learning models tend to keep learning and improving when more data is fed to them. While there are many different types of machine-learning approaches, Joseph is interested in one called natural language processing . You are probably pretty familiar with an example of natural language processing at work – Google Translate! The model that Joseph is building is in fact not too dissimilar from Google Translate, or at least the idea behind it; except that instead of taking English and translating it into Spanish, Joseph’s model is taking RNA and translating (or not translating) it into a protein. In Joseph’s own words, “We’re going through this whole rigamarole [aka his PhD] to understand how the ins [RNA & ribosomes] create the outs [proteins].”.
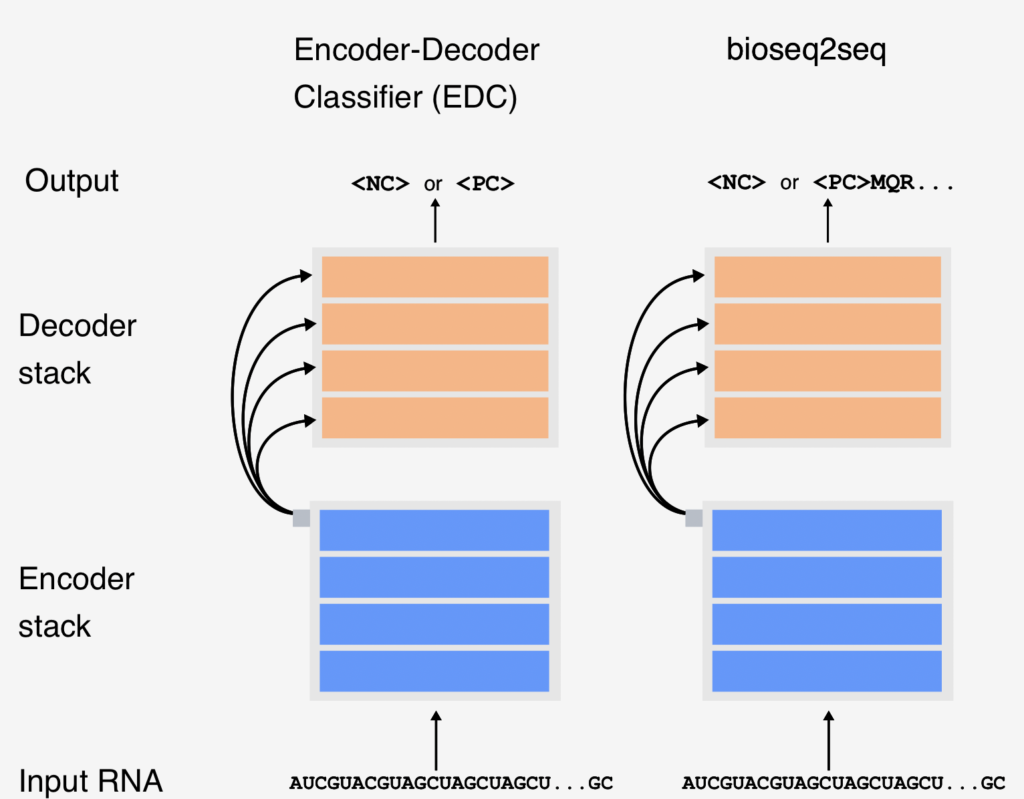
But it is not as easy as it sounds. There are a lot of complexities to the work because the thing that makes machine learning so powerful is that the exact complexities that gives these models the power that they have, also makes it hard to interpret why the model is doing what it is doing. Even a highly performing machine learning model may not capture the exact biological rules that govern translation, but successfully interpreting its learned patterns can help in formulating testable hypotheses about this fundamental life process.
To hear more about how Joseph is building this model, how it is going, and what brought him to OSU, listen to the podcast episode! Also, you can check out Joseph’s personal website to learn more about him & his work!
