The CGRB’s biocomputing infrastructure was highlighted in AMD CEO Lisa Su’s keynote speech at the Consumer and Electronics Show (CES) in Las Vegas on January 9, 2019. Watch below:
Using GPU’s to Classify Oceans of Data
This blog post was originally published on September 10, 2018 and written by Christopher M. Sullivan, Assistant Director for Biocomputing. Read the whole article here.
The Oregon State University’s Center for Genome Research and Biocomputing (CGRB) and the Plankton Ecology Lab at OSU Hatfield have been collaborating in implementing an image processing pipeline to automate the classification of in situ images of plankton: microscopic organisms at the base of the food web in the world’s oceans and freshwater ecosystems. The imagery collection from a 10-day cruise typically contains approximately 80 TB worth of video, which, in some cases, may convert into image data yielding several billions of segments representing individual plankton and particles that need to be identified; a near impossible task to carry out manually by human experts. While we have a fully functional Convolutional Neural Net (CNN) algorithm that does an excellent job at predicting the identity of the plankton organisms or particles, we have been limited by GPU computational capabilities. We started working with PCI bus based Tesla K40 and K80 GPUs, which were good enough to manage millions of segments. However, when it came to billions of segments, it became a near insurmountable challenge.

R-tips: A Table Makeover with DT
R’s default print function for data frames and matrices is not an effective way to display the contents, especially in a html report. RStudio created a R package, DT, that is an interface to the JavaScript library DataTable. DT allows users to have interactive tables that includes searching, sorting, filtering and exporting! I routinely use these tables in my analysis reports.
Install the DT package from cran
First, one must install and load the DT package. Open up RStudio and run the following commands to install and load the DT package:
# Install the DT package
install.packages("DT")
# Load the DT package
library(DT)
Example Table
The print function is not the most effective was to display a table in an HTML R Markdown report.
print(head(mtcars))
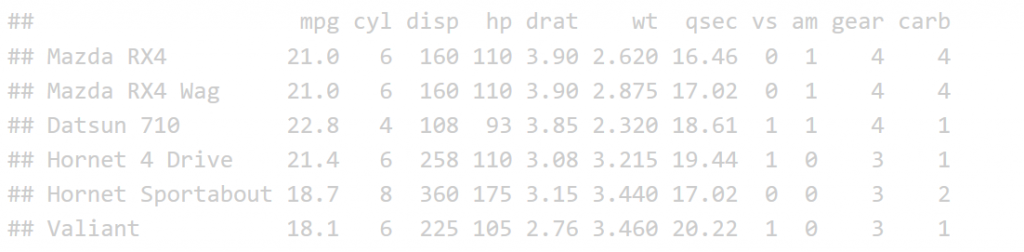
Now let’s look at the datatable function for comparison. The input to the datatable function is a data frame or matrix. Let’s make a table with the preloaded iris data that’s in a data.frame. The basic call is DT::datatable(iris) but in our example I’ve added the filter option to the top of the table, and limited the number of entries to 5 per table. See code and table features below:
datatable(iris, filter = "top",
options = list(pageLength = 5))
A screen shot of the output looks like:
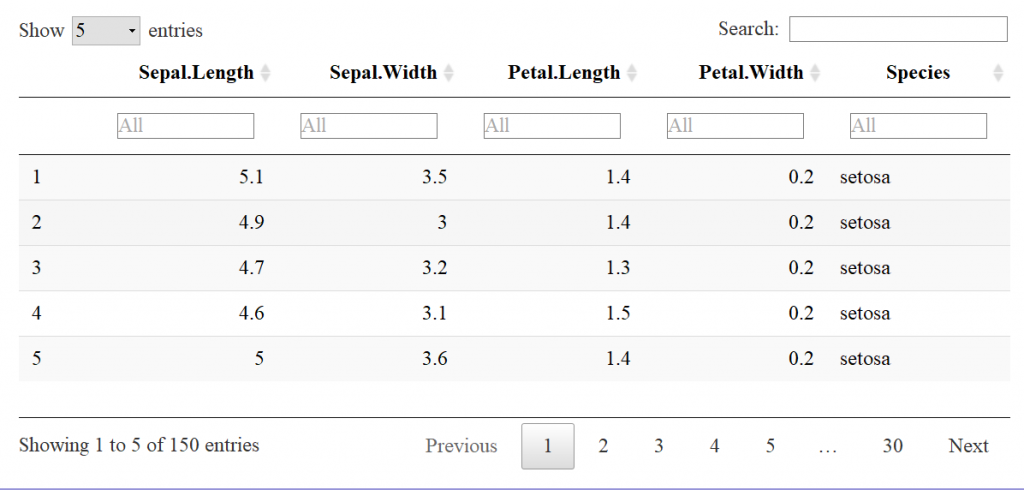
Already, the readability is much better than the base r function print. This is a JavaScript based table, stored in a HTML widget, so a flat image doesn’t convey all of the interactive features.
Features
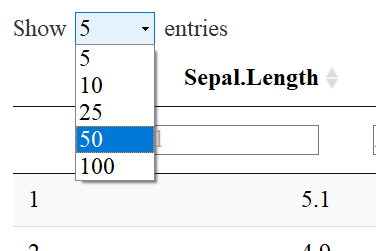
NUMBER OF ENTRIES TO DISPLAY
You’ll notice that there is a drop down menu that says: “Show 5 entries”. The default is 10, but I specified 5 as default with the code pageLength=5. One may select the number of entries to show by using the drop down menu like so:
SEARCH BAR
The widget also includes a search bar on the top right corner which can be very useful when interactively exploring data. Note at the bottom of the table it shows you how many entries (rows) were found and are being displayed.


SORT COLUMNS
Notice that to the right of each column name are two arrows: ![]() One may sort by ascending or descending order and the direction of the blue arrow indicates by which direction you sorted the column.
One may sort by ascending or descending order and the direction of the blue arrow indicates by which direction you sorted the column.

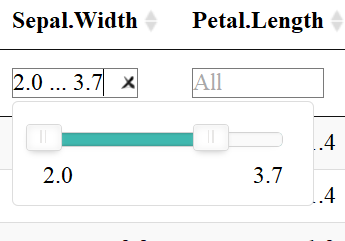
FILTER COLUMNS
The datatable function also allows users to filter each column depending on the datatype: filter numeric columns with a slider & filter columns of class factor with a drop down menu. One must add the filter = "top" (or bottom, etc.) to the code to enable this feature.
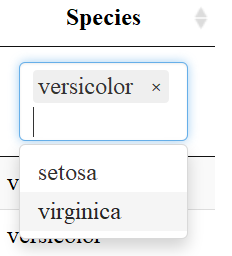
Export Data
Another useful aspect of the datatable function is the “Buttons” extension. This enables users to copy the table, save as a csv, excel or PDF file, or print the table. The table “remembers” what you’ve changed so far—so if you sort by Sepal Length, filter pedal width to > 1 and select species “versicolor” the copied/saved table will have these same restrictions.
datatable(iris,
extensions = 'Buttons',
options = list(dom = 'Bfrtip',
buttons = c('copy', 'csv', 'excel', 'pdf', 'print'))
The above code adds “buttons” to the top of the table like so:
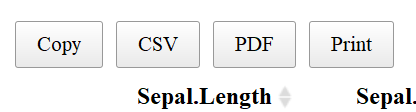
If one clicks “copy”, the table will be copied to your clipboard, “CSV” or “PDF” will save the table to the give file type, and “print” will bring put the table into a print friendly format and will bring up the print dialog box.
Links and Color
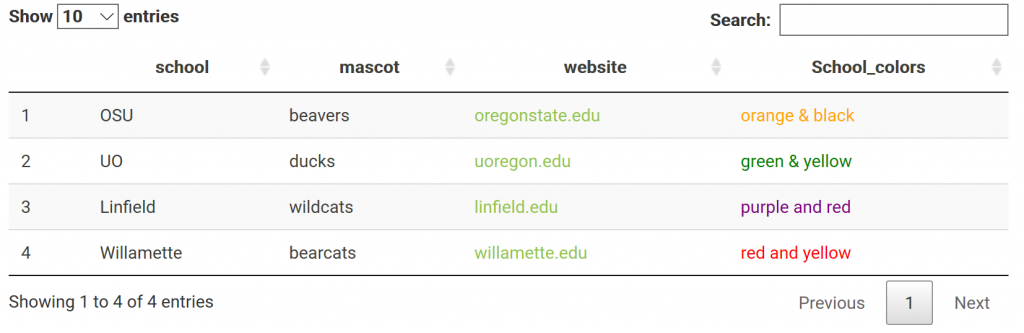
One may also have links in their table. Say you made a data frame with links you want to work in your html report. For example: a data frame of variants w/ links to their position in a genome browser. This is done through not escaping content in the table, specifically the column with the links. The links are made with html and must not be escaped to show up. This applies to other html as well; including color. For me, it was confusing that I had to not escape the html columns. Got it completely backwards the first time I tried it. NOTE: > got replaced with “& gt;” (with no spaces) when it is rendered on the blog… Need to find a fix!
# Make dataframe
df.link <- data.frame(school=c("OSU", "UO", "Linfield", "Willamette"),
mascot=c("beavers", "ducks", "wildcats", "bearcats"),
website=c('<a href="http://oregonstate.edu/">oregonstate.edu</a>',
'<a href="https://www.uoregon.edu/">uoregon.edu</a>',
'<a href="https://www.linfield.edu/">linfield.edu</a>',
'<a href="https://www.willamette.edu/">willamette.edu</a>'),
School_colors=c('<span style="color:orange">orange & black</span>',
'<span style="color:green">green & yellow</span>',
'<span style="color:purple">purple and red</span>',
'<span style="color:red">red and yellow</span>'))
# When the html columns, 3 & 4, are not escaped, it works!
datatable(df.link, escape = c(1,2,3))
Column Visibility
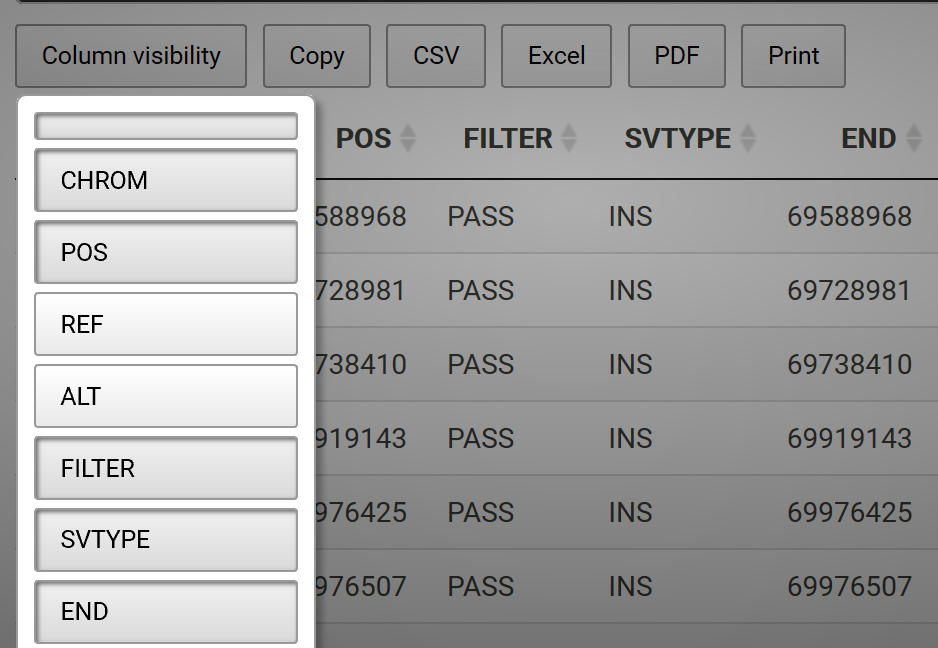
One may also hide columns from visibility and add a button to add the column back interactively. For example, say we have a data frame called sv.all.i.in. We can hide columns 3 and 4, which are long sequences and disrupt the readability of the table, with the following code:
datatable(sv.all.i.in, extensions = 'Buttons',
options = list(dom = 'Bfrtip', columnDefs = list(list(visible=FALSE, targets=c(3,4))),
buttons = list(I('colvis'),c('copy', 'csv', 'excel', 'pdf', 'print'))))
Learn More
There are many more useful features that you can add to your datatable! Learn more here: https://rstudio.github.io/DT/
2019 Fall Conference registration now open
The CGRB 2019 Fall conference registration is now open! Please join us for our annual event this September for informative talks, posters and a reception. This year the Fall Conference will also include lighting talks.
Important Information:
- When: Friday, September 20, 2019
- Where: CH2M Hill Alumni Center – Oregon State University
- Registration: here
- Registration fee: $25 (includes lunch and social hour)
- NOTE: registration fee is waved for:
- Undergrads presenting their research poster
- Lightning talk presenters
- Deadlines:
- Lightning Talk Submission: August 15th
- Poster Registration: September 7th
- Conference Registration: September 13th (registration fee increases to $35 after Sept 13)
Featured Speakers

Ed Kelly
Associate Professor
School of Pharmacy
University of Washington

Daniel Liefwalker
Research Assistant Professor
Molecular and Medical Genetics
Oregon Health and Science University

Doris Taylor
Director
Regenerative Medicine Research
Texas Heart Institute
Conference Agenda
| Time | |
| 8:00-8:50 | Registration & refreshments (Poster & sponsor setup) |
| 8:50-9:15 | Brett Tyler, Director, CGRB Introduction, CGRB update |
| Morning Session | Hosted By Jaga Giebultowicz |
| 9:15 – 9:40 | Andrew Annalora, Environmental and Molecular Toxicology Exploring Splice Variant Biology in Nuclear Receptor and Cytochrome P450 Genes |
| 9:40 – 10:30 | Ed Kelly, University of Washington Organs on a Chip – Chips in Space |
| 10:30 – 10:55 | Break (Poster and Sponsor displays) |
| 10:55 – 11:35 | Morning Lightning Talks (8 talks) – moderated by Jeff Anderson |
| 11:35– 12:00 | Felipe Barreto, Integrative Biology Genomics in the Tidepool: Functional and Population Genetics of Adaptation and Speciation in a Tiny Crustacean |
| 12:00 – 12:25 | Kevin Brown, College of Pharmacy Adventures in Complex Systems |
| 12:25 – 1:25 | Lunch (Poster and Sponsor displays) |
| Afternoon Session | Hosted By Craig Marcus |
| 1:25 – 1:50 | Afua Nyarko, Biochemistry and Biophysics Selectivity and Specificity in Cancer Regulatory Proteins |
| 1:50 – 2:40 | Daniel Liefwalker, Oregon Health and Science University Therapeutic strategies targeting c-MYC |
| 2:40 – 3:20 | Afternoon Lightning Talks (8 talks) – Moderated By Viviana Perez |
| 3:20 – 3:45 | Break 25 mins (Poster and Sponsor displays) |
| 3:45 – 4:10 | Morgan Giers, Chemical, Biological, and Environmental Engineering Regenerating the Intervertebral Disc: Developing Effective Therapies in a Nutrient Limited Environment |
| 4:10 – 5:00 | Doris Taylor, Texas Heart Institute Building Solutions for Heart Disease: A 2019 Update |
| 5:00 – 7:30 | Poster Session / Reception, Sponsor Displays |
Call for posters!
Invitation to present a Poster at the 2019 CGRB FALL CONFERENCE (Sept 20, 2019)
Students, Post Docs, Research Staff and Research Faculty are invited to present their research as a Poster. Presenters are strongly encouraged but not required to consider utilizing a revolutionary new trend in poster format: https://twitter.com/mikemorrison


https://osf.io/ef53g
(Posters in any format displayed at recent meetings are also welcome)
Prizes for Best Posters: $100 (Undergraduate, Graduate and Post Doc Categories).
All fields and research topics welcome. To submit a Poster, please navigate to https://beav.es/ZPd
DEADLINE Sept. 7, 2019.

Call for lightning talks!
Invitation to present a Lightning Talk at the 2019 CGRB FALL CONFERENCE (Sept. 20, 2019)
https://cgrb.oregonstate.edu/fall-conference
Students, Post Docs, Research Staff (FRA, Res. Associates, etc.), and Research Faculty are invited to present their research as a 5-minute Lightning Talk at the annual CGRB Fall Conference, Friday Sept. 20, 2019.
First Prize for Best Lightning Talk = $100
Conference Registration Fee is waivedfor all Lightning Talk Presenters
All fields and research topics welcome. To submit a lightning talk: Please navigate to beav.es/ZPA

Talks are limited to 5 minutes and 5 slides maximum. Please Submit no Later Than: August 15, 2019.
Talks will be selected by the Program Committee and Presenters notified by Aug. 31, 2019.
Committee Members
Thank you to our 2019 Fall Conference Committee:
Jaga Giebultowicz, Department of Integrative Biology
Craig Marcus, Environmental and Molecular Toxicology
Jeff Anderson, Department of Botany and Plant Pathology
Viviana Perez, Department of Biochemistry and Biophysics
Fall Conference 2018
Friday, October 12, 2018
Poster Award Winners
Undergraduate student: Michaela Buchanan, Computer Sci/Biochemistry & Biophysics Using GPUs to Classify Oceans of Data
Graduate student: Courtney Armour, MCB A Metagenomic Meta-Analysis Reveals Functional Signatures of Health and Disease in the Human Gut Microbiome
Postdoc/Trainer: T. William O’Neill, Biomedical Sciences Creating 3-Dimensional Models from Serial Histologic Sections Using Open-Source Software
| 8:00 | Registration & refreshments (Poster & sponsor setup) |
| 9:00 | Brett Tyler, Director, CGRB Introduction |
| 9:30 | Helen Poynton, University of Massachusetts The Toxicogenome of Hyalella Azteca, building genomic resources for healthy ecosystems |
| 10:20 | Break (Poster & sponsor setup / displays) |
| 10:45 | Gerrad Jones, Biological & Ecological Engineering Environmental Change Drives Global Soil Element Fluxes |
| 11:15 | James Watson, College of Earth, Ocean & Atmospheric Science Manifold Learning of Multi-scale Dynamics in Complex Systems |
| 11:45 | Joe Christison, Office of Commercialization & Corporate Development Patenting and Commercializing University Discoveries |
| 12:10 | Lunch (Poster and Sponsor setups / displays) |
| 1:15 | Daniel Coleman, OHSU Targeting BET Bromodomain Proteins in Castration-Resistant Prostate Cancer |
| 2:10 | Maria Franco, Biochemistry & Biophysics Role of Redox Signaling in Tumor Metabolic Reprogramming |
| 2:40 | James Strother, Integrative Biology Elucidation of neural circuit structures through imaging, optogenetics, and behavior |
| 3:05 | Break |
| 3:30 | Maude David, Depts of Microbiology & Pharmacy Children with autism and their typically-developing siblings differ in exact sequence variants and predicted functions of stool-associated microbes |
| 4:00 | Vladimir Shulaev, University of North Texas Neuroprotective effect of 17β-Estradiol (E2) on rat brain |
| 4:50 | Closing remarks |
| 5:00 – 7:30 | Poster Session/Reception, Sponsor Displays |
Thank you to our conference committee:
Jared LeBoldus, Assistant Professor, Botany & Plant Pathology
Claudia Maier, Professor, Chemistry
Natalia Shulzhenko, Assistant Professor, Vet Biomedical Science
CGRB Seminar Series 2018-2019
| Fall Term 2018 | |
| September 26, 2018 | Andrew Cato Karlsruhe Institutue of Technology, Germany Development of Bag-1L as a Therapeutic Target in Androgen Receptor-Dependent Prostate Cancer Host: Siva Kolluri |
| October 10, 2018 | Samara Reck-Peterson UC San Diego Molecular mechanisms of microtubule-based motors: how teams of motors work Host: Michael Freitag |
| October 24, 2018 | Siobhan Brady UC Davis From networks to switches: systems approaches to unravel the control of biological processes necessary for plant life Host: John Fowler/Molly Megraw |
| November 7, 2018 | Fitnat Yildiz UC Santa Clara Mechanisms and regulation of biofilm formation in Vibrio cholerae Host: Claudia Hase |
| Winter Term 2019 | |
| Feruary 8, 2019 | William Hersh OHSU Three amazing ideas about microbial biogeography that will blow your mind Host: Denise Hynes |
| March 6, 2019 | Nadja Cech University of North Carolina – Greensboro Anti-Virulence Strategies Against Drug Resistant Superbugs Host: Sandra Loesgen |
| Spring Term 2019 | |
| April 3, 2019 | Chris Marx University of Idaho Phenotypic heterogeneity between genetically identical cells permits growth with letal levels of formaldehyde stress Host: Patrick de Leenheer |
| April 17,. 2019 | Melissa Haendel OHSU/OSU The Yellow Brick Road of Open Science: Barriers to bringing data to its highest valued use Host: Brett Tyler |
| May 1, 2019 | Kristine Alpi OHSU Host: Melissa Haendel |
| May 15, 2019 | Lucia Carbone OHSU Host: Tom Sharpton |
| May 29, 2019 | Andrew Kern University of Oregon Putting the HAL in Haldane: leveraging supervised machine learning for population genetics Host: Aaron Liston |
Spring Conference 2018
Friday, April 27, 2018
Conference Award Winners
Lightning Talk: Ellie Bors, Population genomics of rapid range expansion during marine invasions
Poster: Lilian Padgitt-Cobb, Toward a phased, diploid assembly of the hop genome
Program
| 8:00 – 9:00 | Registration & refreshments (Poster & lightning talk slides setup) |
| 9:00 – 9:10 | Chris Sullivan, Assistant Director for Biocomputing, CGRB Opening Remarks |
| Moderator: Duo Jiang, Assistant Professor, Statistics | |
| 9:10 – 10:00 | Haiyan Huang, UC Berkeley Inferring Gene Interactions and Functional Modules Beyond Standard Statistical Models |
| 10:00 – 10:25 | Break,time for poster setup |
| 10:25 – 10:50 | David Hendrix, Assistant Professor, Biochemistry & Biophysics, EECS Large-scale Automated Annotation and Analysis of RNA Secondary Structure |
| 10:50 – 12:15 | Lightning talks Moderator: Molly Megraw, Assistant Professor, Botany & Plant Pathology |
| 12:15 – 1:20 | Lunch and Poster Session (Poster & Lightning Talk slides judged) Poster & lightning talk ballot envelopes will be picked up for counting right after lunch |
| Moderator: Rebecca Hutchinson, Assistant Professor, Fisheries & Wildlife, EECS | |
| 1:20 – 1:45 | Susan Tilton, Assistant Professor, Environmental & Molecular Toxicology A 3D Vitro Respiratory Model for Predicting Chemical Toxicity |
| 1:45 – 2:10 | Stephen Ramsey, Assistant Professor, Biomedical Sciences, EECS Machine-Learning for Identifying Functional Noncoding SNPs |
| 2:10 – 2:35 | Clinton Epps, Associate Professor, Fisheries & Wildlife, CAS Genetic applications in the Conservation and Management of Bighorn Sheep |
| 2:35 – 3:00 | Break |
| 3:00 – 3:50 | Fei Fang, Carnegie Mellon Machine Learning and Game Theory for Biodiversity Conservation |
| 3:50 – 4:00 | Vrushali Bokil, Associate Professor, Mathematics Awards and Closing Remarks |
Thank you to our conference committee:
Vrushali Bokil – Associate Professor, Mathematics
Rebecca Hutchinson – Assistant Professor, EECS, Fisheries & Wildlife
Duo Jiang – Assistant Professor, Statistics
Molly Megraw – Assistant Professor, Botany & Plant Pathology
Fall Conference 2017
Friday, September 8, 2017
Poster Award Winners
Postdoc/Trainee: Fadi El-Rami, Subcellular proteomics of the 2016 WHO Neisseria gonorrhoeae reference strains for identification of vaccine candidates and antibiotic resistance signatures
Grad student: Aayushi Manchanda, Loss of otoferlin alters the transcriptome profile and ribbon synapse architecture of sensory hair cells
Undergrad student: Ryan Kitchen, CASSA: The new fast GPU DNA Sequence Aligner
Program
| 8:00 | Registration & refreshments (Poster & sponsor setup) |
| 9:00 | Brett Tyler, Director, CGRB Introduction, CGRB update |
| Moderator: Aleksandra Sikora, Pharmaceutical Sciences | |
| 9:25 | Caroline Attardo Genco, Tufts University Sex as a variable: Distinct Neisseria gonorrhoeae gene expression signatures expressed during mucosal infection in men and women |
| 10:10 | Break (Poster & sponsor setup / displays) |
| 10:30 | Nathan Campbell, Columbia River Inter-Tribal Fish Commission Genotyping in Thousands by sequencing: A low cost, high-throughput, targeted SNP genotyping method |
| 11:15 | Olena Taratula, College of Pharmacy A Tumor-Activatable Theranostic Nanomedicine Platform for NIR Fluorescence-Guided Surgery and Phototherapy |
| 11:40 | Posy Busby, Botany & Plant Pathology Host genetic structuring of leaf endophyte communities in Populus and Maize |
| 12:05 | Lunch (Poster and Sponsor setups / displays) |
| Moderator: Brent Kronmiller, CGRB | |
| 1:20 | Melissa Haendel, OHSU Crossing the chasm of semantic despair: ontologies to unify genomics and phenomics |
| 2:05 | Brett Tyler, Botany & Plant Pathology Does the pathogen always win – the co-evolutionary struggle between soybean and its oomycete pathogen Phytophthora sojae |
| 2:30 | Molly Burke, Integrative Biology Genomic lessons from laboratory evolution experiments |
| 2:55 | Break |
| 3:15 | Marian Waterman, UC Irvine WNT signaling and tumor heterogeneity |
| 4:00 | Maria Clara Franco, Biochemistry & Biophysics The Two Faces of Nitrated Proteins: Neurodegeneration and Cancer |
| 4:25 | David Myrold, Crop & Soil Science Glimpses into the microbial communities of Oregon soils |
| 4:50 | Janine Trempy, Associate Vice Provost for Academic Affairs Closing remarks |
| 5:00 – 7:30 | Poster Session/Reception, Sponsor Displays |
Thank you to our conference committee:
Pankaj Jaiswal, Botany & Plant Pathology
Ryan Mehl, Biochemistry & Biophysics
Aleksandra Sikora, Pharmaceutical Sciences
CGRB Seminar Series 2017-2018
| Fall Term 2017 | |
| September 27, 2017 | Geoffrey Wahl The Salk Institute Understanding Intra-tumoral heterogeneity using a developmental lens Host: Arup Indra |
| October 11, 2017 | David Thomas Biochemistry, Molecular Biology and Biophysics University of Minnesota Muscle Protein Structural Dynamics and Therapeutic Discovery Host: Weihong Qiu |
| October 25, 2017 Cosponsored by the Departments of Microbiology and Chemistry ROOM CHANGE **Withycombe 109** | Paul Jensen Center for Marine Biotechnology & Biomedicine UC San Diego Ecological and evolutionary perspectives on secondary metabolism in marine actinobacteria Host: Sandra Loesgen |
| November 8, 2017 | Brenna Henn Stony Brook University What do we gain by studying African genomes? : Examples from human evolution Host: Thomas Sharpton |
| December 6, 2017 | Pat Schloss University of Michigan Understanding Disease through the Lens of the Microbiome Host: Thomas Sharpton |
| Winter Term 2018 | |
| February 7, 2018 | Scott Landfear Dept of Molecular Microbiology & Immunology OHSU Critical Roles for Glucose Transporters in Parasitic Protozoa: From Cell Biology to Drug Discovery Host: Sandra Loesgen |
| February 21, 2018 | Jason Slot Fungal Evolutionary Genomics The Ohio State University Investigating fungal chemical ecology with evolutionary genomics Host: Michael Freitag |
| March 7, 2018 | Rosalie Sears Molecular and Medical Genetics OHSU Modeling and targeting intra-tumor phenotypic heterogeneity and cellular plasticity Host: Siva Kolluri |
| Spring Term 2018 | |
| April 4, 2018 | John Taylor UC Berkeley The species problem for fungi in the era of genomics Host: Nik Grünwald |
| April 18, 2018 | Catherine Royer Biological Sciences Rensselaer Polytechnic Institute Pressure-based mapping of protein conformational landscapes Host: Elisar Barbar |
| May 2, 2018 | Maret Traber Linus Pauling Institute Oregon State University Ferroptosis, Mechanism of Cell Death in Vitamin E Deficiency During Embryogenesis? |
| May 16, 2018 | Steve Reichow Chemistry Portland State University Native Lens Gap Junctions Visualized at Near-Atomic Resolution by CryoEM Host: Elisar Barbar |
| May 29, 2018 NOTE: Date change Tuesday, May 29 3:30-4:30 ALS 4001 | Rebecca Fry Environmental Sciences and Engineering UNC Chapel Hill The placental epigenome as a driver of early and later life health effects Host: Molly Kile |
Industry Symposium Program
Wednesday, June 21, 2017
Program provisional
| 8:00 | Registration & refreshments (Poster setup) |
| 8:30 | Brian Wall, Assistant Vice President, Research, Commercialization & Industry Partnerships, Oregon State University Introduction |
| Session 1 | Moderator: Joseph McGuire, OSU; Monique Lajeunesse, OSU |
| 8:40 | Joseph Beckman, Distinguished Professor, Environmental & Molecular Toxicology, Oregon State University Towards curing ALS (Lou Gehrig’s disease) and other neurodegenerative diseases. |
| 9:00 | Laura Heiser, Assistant Professor, Biomedical Engineering, Oregon Health Science University |
| 9:20 | John Bial, CEO, Yecuris Commercial Translation, taking ideas from theory to practice |
| 9:40 | Kristina Young, Earle A. Chiles Research Institute Translational Research: Rational and Relevant Science |
| 10:00 | Discussion |
| 10:20 | Coffee Break |
| Session 2 | Moderator: Jeff Chang, OSU; Kristof Torkenczy, OHSU |
| 10:50 | Hilary Ely, Life Sciences Applications R&D Engineer, HP Inc. Advancing HP Beyond Ink: Applying HP’s technology to life sciences and healthcare |
| 11:10 | Robyn Tanguay, Distinguished Professor, Environmental & Molecular Toxicology, Oregon State University Integrating the power of the zebrafish model with automation to accelerate scientific discoveries |
| 11:30 | Sandra Rugonyi, Associate Professor, Biomedical Engineering, Oregon Health Science University Predicting risks and outcomes in cardiovascular disease. Progress and challenges |
| 11:50 | Yigit Menguc, Assitant Professor, Mechanical Engineering, Oregon State University Soft Robotics combining Soft Materials, Mechanisms, and Manufacturing |
| 12:10 | Discussion |
| 12:30 | Lunch and poster session |
| Session 3 | Moderator: Michaele Armstrong, Oregon Bioscience Association; Sigrid Noreng, OHSU |
| 1:30 | Kyle Ellrott, Assistant Professor, Biomedical Engineering, Oregon Health Science University Big Data and Cancer Genomics |
| 1:50 | Elain Fu, Assistant Professor, Bioengineering, Oregon State University Paper-based testing for precision health |
| 2:10 | Matthew Ryder, Research & Development Surface Biochemist, Qorvo Qorvo Biosensors |
| 2:30 | Summer Gibbs, Assistant Professor, Biomedical Engineering, Oregon Health Science University |
| 2:50 | Discussion |
| 3:10 | Coffee Break |
| Session 4 | Moderator: Summer Gibbs, OHSU; Ward Shalash, OSU |
| 3:40 | Karl Schilke, Assistant Professor, Bioengineering, Oregon State University Bionanoparticles: A versatile platform technology for personalized medicine and bioprocessing |
| 4:00 | Brian Maloney, Chemical Engineer, Patheon Patheon Oregon Site Overview |
| 4:20 | Greg Herman, Professor, Chemical Engineering, Oregon State University See Through Sensors |
| 4:40 | Discussion |
| 5:00 | Brett Tyler, Director, Center for Genome Research & Biocomputing, Oregon State University Closing |
| 5:10 – 7:00 | Poster Session and Networking Reception (cash bar) |