By Solène Derville, Postdoc, OSU Department of Fisheries, Wildlife, and Conservation Science, Geospatial Ecology of Marine Megafauna Lab
About 10 months have passed since I started working on OPAL, a project that aims to identify the co-occurrence between whales and fishing effort in Oregon to reduce entanglement risk. During this period, you would be surprised to know how little ecology I have actually done and how much time has been devoted to data processing! I compiled several million GPS trackline positions, processed hundreds of marine mammal observations, wrote several thousand lines of R code, downloaded and extracted a couple Gb of environmental data… before finally reaching the modeling phase of the OPAL project. And with it, finally comes the time to look more closely at the ecology and behavior of my species of interest. While the previous steps of the project were pretty much devoid of ecological reasoning, the literature homework now comes in handy to guide my choices regarding habitat use models, such as selecting environmental predictors of whale occurrence, deciding on what seasons should be modeled, and choosing the spatio-temporal scale at which the data should be aggregated.
Whale diversity on the US west coast
The productive waters off the US west coast host a great diversity of cetaceans. Eight species of baleen whales are reported to occur there by NOAA fisheries: blue whales, Bryde’s whales, fin whales, gray whales, humpback whales, minke whales, North Pacific right whales and sei whales. Among them, no less than five are listed as Endangered under the Endangered Species Act. Whether they are only passing by or spending months feeding in the region, the timing and location where these animals are observed varies greatly by species and by population.
During the 113 hours of aerial survey effort and 264 hours of boat-based search conducted for the OPAL project, 563 groups of baleen whales have been observed to-date (up to mid-May 2021 to be exact… more data coming soon!). Among the observations where animals could be identified to the species level, humpback whales are preponderant, as they represent about half of the whale groups observed (n = 293). Blue (n = 41) and gray whales (n = 46) come next, the latter being observed in more nearshore waters. Finally, a few fin whale groups were observed (n = 28). The other baleen whale species reported by NOAA in the US west coast species list were very rarely or not observed at all during OPAL surveys.
The OPAL aerial surveys conducted in partnership with the United States Coast Guard (USCG) were specifically designed to study whales occurring on the continental shelf along the coast of Oregon. Hence, most of this survey effort is located in waters from 800 m to 30 m deep, which may explain the relatively low number of gray whales detected. Indeed, gray whales observed in Oregon may either be migrating along the coast to and from their breeding grounds in Baja California, or be part of the small Pacific Coast Feeding Group that forage in Oregon nearshore and shallow waters during the summer. This group of whales is one the main GEMM lab’s research focus, being at the core of no less than three ongoing research projects: AMBER, GRANITE, and TOPAZ.
So today, let’s turn our eyes to the sea horizon and talk about some other members of the baleen whale community: rorquals. Conveniently, the three species of baleen whales (gray whales aside) most commonly observed during OPAL surveys are all part of the rorqual family, a.k.a Balaenopteridae: humpback whales, blue whales and fin whales (Figure 1). They are morphologically characterized by the pleated throat grooves that allow them to engulf large quantities of food and water, for instance when lunge-feeding. Known cases of hybridization between these three species demonstrate their close relatedness (Jefferson et al., 2021). They all have worldwide distributions and display unequally understood migratory behaviors, seasonally traveling between warm tropical breeding grounds and temperate-polar feeding grounds. They occur in great numbers in productive waters such as the upwelling system of the California Current.
The three accomplices

Humpback whales (Megaptera novaeangliae) are easily differentiated from other rorquals because of their long pectoral fins (up to one third of their body length!), which inspired their scientific name, Megaptera, « big-winged » (Figure 1). Individuals observed in Oregon mostly belong to a mix of two Distinct Population Segments (DPS): the threatened Mexico and endangered Central American DPS. Although humpback whales from different DPS do not show any morphological differences, they are genetically distinct because they have been mating separately in distinct breeding grounds for generations and generations. This genetic differentiation has great implications in terms of conservation since the Central American DPS is recovering at a lesser rate than the Mexican and is therefore subject to different management measures (recovery plan, monitoring plan, designated critical habitats). Humpback whales migrate and feed off the US west coast, with a peak in abundance in the mid to late summer. Compared to other rorquals that are found in the open ocean, humpback whales are mostly observed on the continental shelf (Becker et al., 2019). They are considered to have a relatively generalist diet, as they feed on a mix of krill (Euphausiids) and fishes (e.g. anchovy, sardines) and are capable of switching their feeding behavior depending on relative prey availability (Fleming, Clark, Calambokidis, & Barlow, 2016; Fossette et al., 2017).
Blue whales (Balaenoptera musculus) are the largest animals ever known (max length 33 m, Jefferson et al., 2008), and sadly the most at risk of global extinction among our three species of interest (listed as « endangered » in the IUCN red list). They have a distinctive mottled blue and light gray skin, a slender body and a broad U-shaped head (or as some say « like a gothic arch », Figure 1). Blue whales tend to be open ocean animals, but they regroup seasonally to feed in highly productive nearshore areas such as the Southern California Bight (Becker et al. 2019, Abrahms et al. 2019). Blue whales migrating or feeding along the US west coast belong to the Eastern North Pacific stock and are subject to great research and conservation efforts. Contrary to their other rorqual counterparts, blue whales are quite picky eaters, as they exclusively feed on krill. This difference in diet leads to resource partitioning facilitating rorqual coexistence in the California Current (Fossette et al., 2017). These differences in feeding strategies have important implications for designing predictive models of habitat use.
Fin whales (Balaenoptera physalus) are nicknamed « greyhounds of the sea » due to their exceptional swim speed (max 46 km/h). They are a little smaller than blue whales (max length 27 m, Jefferson, Webber, & Pitman, 2008) but share a similar sleek and streamlined shape. Their coloration is their most distinctive feature: the left lower jaw being mostly dark while the right is white. V-shaped light-gray « chevrons » color their back, behind the head (Figure 1). The California/Oregon/Washington is one of the three stocks recognized in the North Pacific (NOAA Fisheries, 2018). Within this region, there is genetic evidence for a geographic separation north and south of Point Conception, CA (Archer et al., 2013). Like other rorquals, they are migratory, but their seasonal distribution is relatively less well understood as they appear to spend a lot of time in open oceans. For instance, a meta-analysis for the North Pacific found little evidence for fin whales using distinct calving areas (Mizroch, Rice, Zwiefelhofer, Waite, & Perryman, 2009). In the California Current System, satellite tracking has provided great insights into their space-use patterns. In the Southern California Bight, fin whales show year-round residency and seasonal shifts in habitat use as they move further offshore and north during the spring/summer (Scales et al., 2017). The Northern California Current offshore waters appeared to be used during the summer months by the whales tagged in the Southern California Bight. Yet, fin whales are observed year-round in Oregon (NOAA Fisheries, 2018).
Towards predictive models of rorqual distribution
Enough observations have now been collected as part of the OPAL project to be able to model the habitat use of some of these rorqual species. Based on 12 topographic (i.e., depth, slope, distance to canyons) and physical variables (temperature, chlorophyll-a, water column stratification, etc.), I have made my first attempt at predicting seasonal distribution patterns of humpback whales and blue whales in Oregon. These models will be improved in the coming months, with more data pouring in and refined parametrizations, but they already bring insights into the shared habitat use patterns of these species, as well as their specificities.
Across multiple cross-validations of the species-specific models, sea surface temperature, sea surface height and depth were recurrently selected among the most important variables influencing both humpback and blue whale distributions. Predicted densities of blue whales were relatively higher at less than 40 fathoms compared to humpback whales, although both species’ hotspots were located outside this newly implemented seasonal fishing limit (Figure 2). Higher densities were generally predicted off Newport and Port Orford, and north of North Bend.
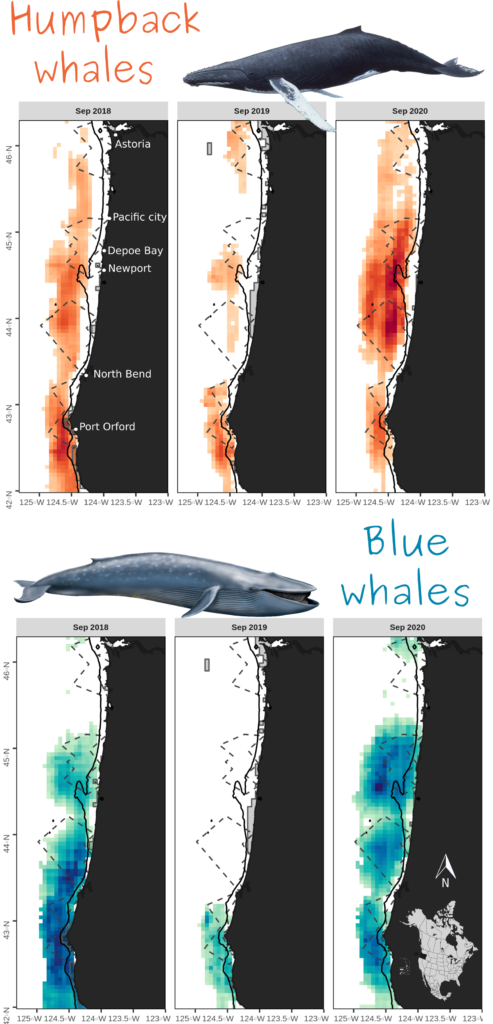
Once our rorqual models are finalized, we will work with our partners at the Oregon Department of Fisheries and Wildlife to overlay predicted whale hotspots with areas of high crab pot densities. This overlap analysis will help us understand the times and places where co-occurrence of suitable whale habitat and fishing activities put whales at risk of entanglement.
References
Becker, E. A., Forney, K. A., Redfern, J. V, Barlow, J., Jacox, M. G., Roberts, J. J., & Palacios, D. M. (2019). Predicting cetacean abundance and distribution in a changing climate. Diversity and Distributions, 25(4), 626–643. https://doi.org/10.1111/ddi.12867
Fleming, A. H., Clark, C. T., Calambokidis, J., & Barlow, J. (2016). Humpback whale diets respond to variance in ocean climate and ecosystem conditions in the California Current. Global Change Biology, 22, 1214–1224. https://doi.org/10.1111/gcb.13171
Fossette, S., Abrahms, B., Hazen, E. L., Bograd, S. J., Zilliacus, K. M., Calambokidis, J., … Croll, D. A. (2017). Resource partitioning facilitates coexistence in sympatric cetaceans in the California Current. Ecology and Evolution, 7, 9085–9097. https://doi.org/10.1002/ece3.3409
Jefferson, T. A., Palacios, D. M., Clambokidis, J., Baker, S. C., Hayslip, C. E., Jones, P. A., … Schulman-Janiger, A. (2021). Sightings and Satellite Tracking of a Blue / Fin Whale Hybrid in its Wintering and Summering Ranges in the Eastern North Pacific. Advances in Oceanography & Marine Biology, 2(4), 1–9. https://doi.org/10.33552/AOMB.2021.02.000545
Jefferson, T. A., Webber, M. A., & Pitman, R. L. (2008). Marine Mammals of the World. A comprehensive guide to their identification. Elsevier, London, UK.
Mizroch, S. A., Rice, D. W., Zwiefelhofer, D., Waite, J., & Perryman, W. L. (2009). Distribution and movements of fin whales in the North Pacific Ocean. Mammal Review, 39(3), 193–227. https://doi.org/10.1111/j.1365-2907.2009.00147.x
NOAA Fisheries. (2018). Fin whale stock assessment report ( Balaenoptera physalus physalus ): California / Oregon / Washington Stock.
Scales, K. L., Schorr, G. S., Hazen, E. L., Bograd, S. J., Miller, P. I., Andrews, R. D., … Falcone, E. A. (2017). Should I stay or should I go? Modelling year-round habitat suitability and drivers of residency for fin whales in the California Current. Diversity and Distributions, 23(10), 1204–1215. https://doi.org/10.1111/ddi.12611